 |
 |
 |
 |
 |
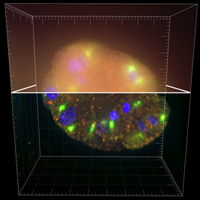
Deconvolution is one of the most common image-reconstruction tasks that arise in 3D fluorescence microscopy. The aim of this challenge is to benchmark existing deconvolution algorithms and to stimulate the community to look for novel, global and practical approaches to this problem.
The challenge will be divided into two stages: a training phase and a competition (testing) phase. It will primarily be based on realistic-looking synthetic data sets representing various sub-cellular structures. In addition it will rely on a number of common and advanced performance metrics to objectively assess the quality of the results.
In the framework of this challenge we provide both datasets and code.
Different data sets have been released for each stage of the challenge.
The link to the evaluation-stage data will be sent to registered participants by e-mail.
Note that the X-Y resolution of these data sets is 80 nm, while the Z resolution is 180 nm.
| Channel | Data stack | PSF |
|---|---|---|
| 0 |

|
|
| 1 |

|
|
| 2 |

|
|
| 3 |
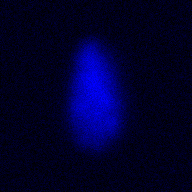
|
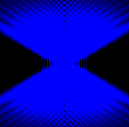
|
The goal of the training stage is to allow the participants to familiarize themselves with our deconvolution setting, by experimenting with a representative dataset.
The training-stage data comprises 4 channels corresponding to different structures (point sources, filaments, membrane, dense object). Maximum-intensity projections of the data and PSFs are shown in the table below. Note that for channel 1 and 2 special care must be taken regarding the boundary condtions.
For the training stage, we have also decided to disclose the parameters of the noise model. The values of the parameters for each channel are given below. Please also see the forward-model documentation for a detailed description of these parameters.
| Channel | Background Constant (b) | Maximum number of photons per voxel (k) | Standard deviation of Gaussian noise (σ) |
|---|---|---|---|
| 0 | 15.8 | 239.6 | 9.7 |
| 1 | 7.8 | 342.4 | 3.1 |
| 2 | 11.2 | 289.8 | 5.6 |
| 3 | 19.6 | 153.8 | 12.3 |
| Channel | Data stack | PSF |
|---|---|---|
| 0 |
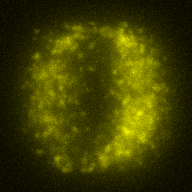
|

|
| 1 |
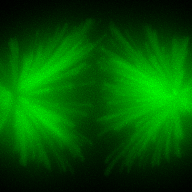
|

|
| 2 |

|
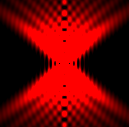
|
| 3 |
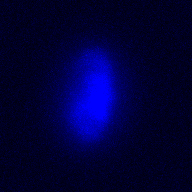
|
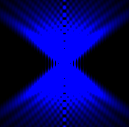
|
In addition to the datasets, we provide registered participants with a collection of Matlab scripts that implement the forward model and some of the performance metrics. Together with the supporting documentation, the aim is to give the participants a precise understanding of our simulation and evaluation framework. In particular, the code allows the participants to 1) generate synthetic micrographs based on their own ground-truth data and 2) to assess and calibrate their deconvolution algorithm.
The registered participants will receive soon a personalized upload link.
January 6, 2014The evaluation stage of the second edition of the challenge has started.
November 6, 2013The official stage of the challenge will last around 2 months.
November/December, 2013The training stage of the second edition of the challenge has started. Follow this link to register and receive e-mail updates.
July 17, 2013