Segmentation and tracking of growing colony of mycobacteria
Spring 2013
Project: 00266
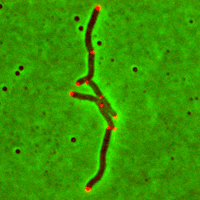
Cell tracking is a recurrent problem in biomedical research: processing datasets on a per-cell basis can potentially unravel cell behaviours that would have escaped notice in a more classical population-based analysis. As large amount of image data that cannot be processed manually are routinely acquired, there is an extensive need for automated solutions.
This project's starting point is a cell segmentation+tracking algorithm that was developed at the BIG in 2011 for the Laboratory of Microbiology and Microsystems (LMIC) based at EPFL. In its current state, the program is not usable as it lacks a good user interface, only allows for limited user interaction, and does not produce quantitative outputs. The goal of this project is to design an interactive tracking tool that would be convenient for biologists. While a large part of this work will be dedicated to coding in Java, an equally important task will be to understand the biologist's need and expectations through regular meetings with the collaborating lab.
The student should ultimately be able to produce a fully working and user-friendly ImageJ plugin to analyze the type of data of interest. We encourage the student to put his/her imagination at work in designing the interface of the program in order to make it generalizable. Other work related to cell tracking could then be done if time permits.
This project's starting point is a cell segmentation+tracking algorithm that was developed at the BIG in 2011 for the Laboratory of Microbiology and Microsystems (LMIC) based at EPFL. In its current state, the program is not usable as it lacks a good user interface, only allows for limited user interaction, and does not produce quantitative outputs. The goal of this project is to design an interactive tracking tool that would be convenient for biologists. While a large part of this work will be dedicated to coding in Java, an equally important task will be to understand the biologist's need and expectations through regular meetings with the collaborating lab.
The student should ultimately be able to produce a fully working and user-friendly ImageJ plugin to analyze the type of data of interest. We encourage the student to put his/her imagination at work in designing the interface of the program in order to make it generalizable. Other work related to cell tracking could then be done if time permits.
- Supervisors
- Virginie Uhlmann, virginie.uhlmann@epfl.ch, 021 693 1136, BM 4.142
- Michael Unser, michael.unser@epfl.ch, 021 693 51 75, BM 4.136
- Daniel Sage