Automatic bone segmentation with active parametric contour in CT images
2022
Master Semester Project
Project: 00424
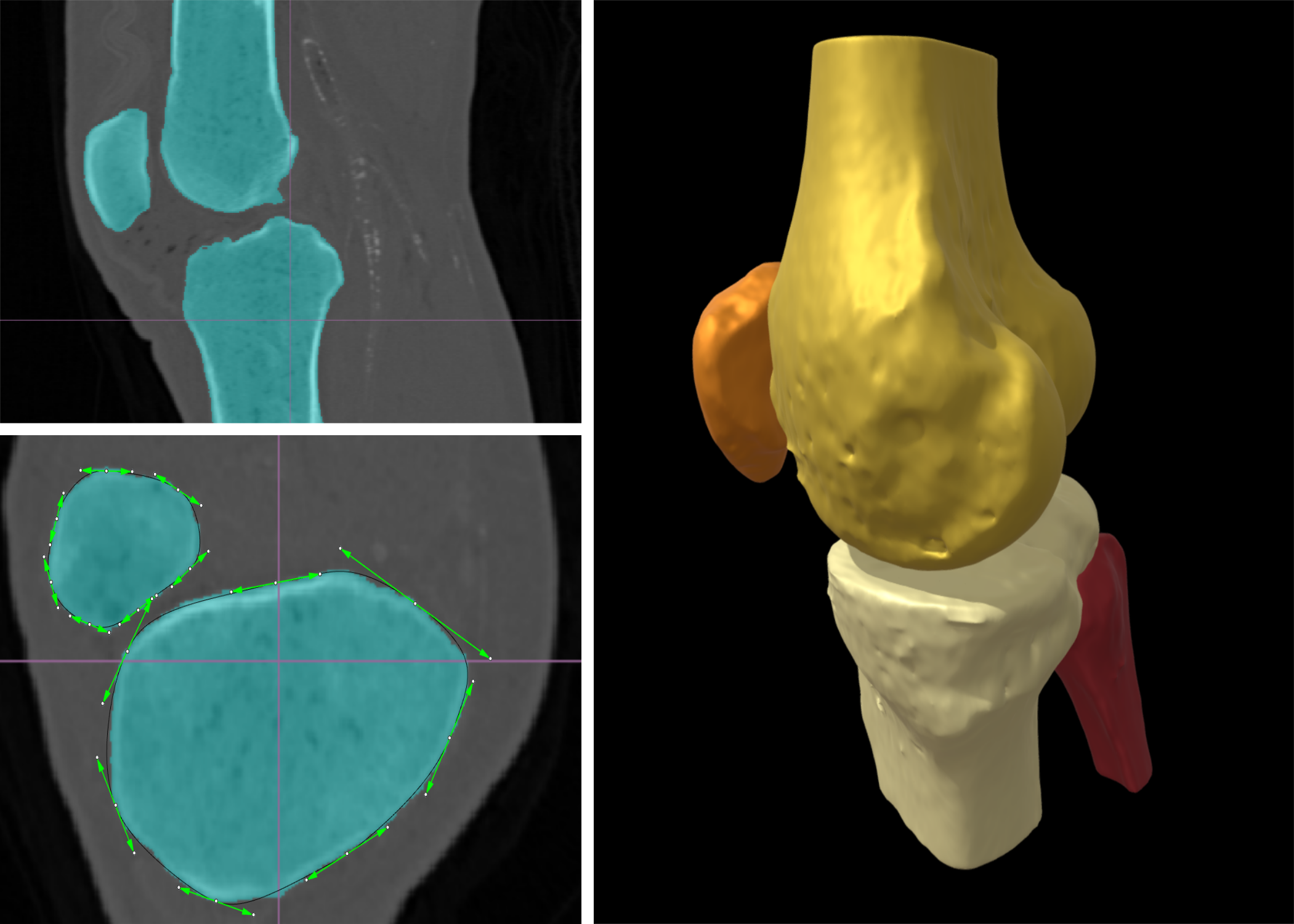
Segmentation and 3D representation of bone structures in CT scans are essential steps for the diagnosis and treatment of skeletal disorders like osteoporosis. Bone has an outer layer of dense protective material (the cortical bone), and an inner light and porous part (the cancellous bone); this complex structure makes manual or semiautomatic segmentation techniques, like thresholding, tedious and impractical for clinical routine.
This project aims to automate the process of bone segmentation in two phases: 1) using convolutional neural networks (with the U-Net architecture) to extract a map of probabilities for bone tissue over the voxelized CT scan; 2) segmenting every slice of the probability map with a parametric curve (active contour), and eventually reconstructing the whole 3D structure with a tensor-product surface.
The project will focus on medical image processing and deep learning techniques using synthetic or experimental data from particular anatomical structures like the knee or vertebral column. It will be co-supervised by a start-up in the field of medical imaging. The student will work with a leading 3D CAD environment, and learn to process and convert the project outputs into standard formats for use in downstream applications like 3D printing and finite element analysis.
The project is in collaboration with a start-up: Mirrakoi SA
This project aims to automate the process of bone segmentation in two phases: 1) using convolutional neural networks (with the U-Net architecture) to extract a map of probabilities for bone tissue over the voxelized CT scan; 2) segmenting every slice of the probability map with a parametric curve (active contour), and eventually reconstructing the whole 3D structure with a tensor-product surface.
The project will focus on medical image processing and deep learning techniques using synthetic or experimental data from particular anatomical structures like the knee or vertebral column. It will be co-supervised by a start-up in the field of medical imaging. The student will work with a leading 3D CAD environment, and learn to process and convert the project outputs into standard formats for use in downstream applications like 3D printing and finite element analysis.
The project is in collaboration with a start-up: Mirrakoi SA
- Supervisors
- Daniel Sage, daniel.sage@epfl.ch, 021 693 51 89, BM 4.135
- Pablo Garcia-Amorena, pablo.garcia-amorena@mirrakoi.com