
Functional morphological evolution of neurones in culture
Anil Swaroop
Section Microtechnique
Student Project
Semester Project, June 2001
Abstract
In many biomedical applications in which alive cells evolution is
studied, it is needed to have a tool allowing the quantitative and not
qualitative following of their evolution, as for example the variation
of the volume of a particular cell. This could be used
to know the reaction of the studied cells when their environment
undergoes changes (pH or physiological serum concentration variations).
Nowadays, the holographic microscopy is a promising method which
allows to acquire images of neurones in culture. A quantitative
description on the evolution of these neurones would be very precious
for the researchers. As the manual measurments on those cells is very
difficult, an automated scheme would be more efficient and faster.
Keywords: snake, active contour, ImageJ
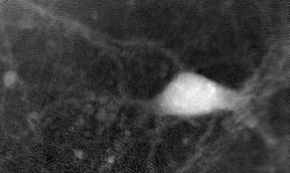
A cell by holographic microscopy
The goal of the project was to realize a plugin for ImageJ which could be use to determine and visualize the evolution of a cell's contour and volume in a sequence of images that are aquired by holographic microscopy. The gray level in images of that kind is linked with the cell thickness. That means one can determine the volume by integrating the gray values inside the cell knowing it's boundary.
Method: Using the SNAKES
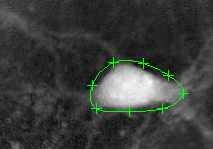
To track the contour of a particular cell, we are using "snakes". A snake is an active contour which is a closed curve (i.e. periodic). The user initializes a first curve around the cell and an algorithme determines a final curve that fits the cell's bondary. For the next image, the final curve of the previous image is used as the initial image.
The plugin
The plugin is used to:
- initialize the snake
- chose the snaking parameters
- run the algorithme
- display the messages / information
- display the results
Results
Initializing the curve:
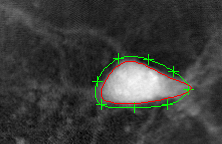
The algorithm finds the cell's boundary:
The evolution is plotted:
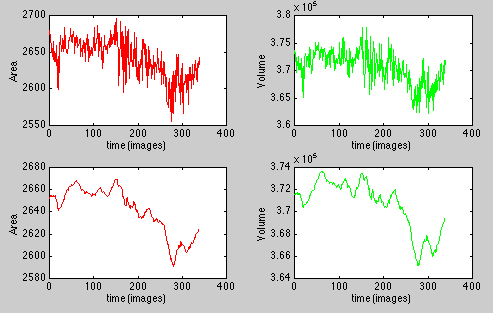
This picture represents the evolution of the area (top left) and the volume (top right) of a 340 images sequence. As there is some noise, the graphs have been smoothed (bottom). This smoothing function is not available in the plugin (it was done with Matlab).
Conclusion
The plugin works and it is possible to follow the evolution (area and volume) of a cell in a sequence of images.