 |
 |
 |
 |
 |
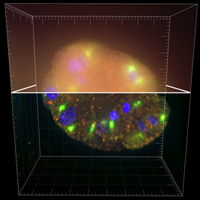
Deconvolution is one of the most common image-reconstruction tasks that arise in 3D fluorescence microscopy. The aim of this challenge is to benchmark existing deconvolution algorithms and to stimulate the community to look for novel, global and practical approaches to this problem.
The challenge will be divided into two stages: a training phase and a competition (testing) phase. It will primarily be based on realistic-looking synthetic data sets representing various sub-cellular structures. In addition it will rely on a number of common and advanced performance metrics to objectively assess the quality of the results.
To read the image stacks we provide the script imreadstack.m. This is
an extension of MATLAB's imread.m and can read an image stack saved in
a graphics file such as .tif. This script accepts a single argument
which should be a string with the full pathname of the file.
We also provide a similar script for writing image stacks: imwritestack.m.
The registered participants will receive soon a personalized upload link.
January 6, 2014The evaluation stage of the second edition of the challenge has started.
November 6, 2013The official stage of the challenge will last around 2 months.
November/December, 2013The training stage of the second edition of the challenge has started. Follow this link to register and receive e-mail updates.
July 17, 2013