Motivation
The main purpose of measuring dendrites is to study the dendrites’ growth. It is characteristic of the neuronal activities, and therefore of the neuron’s capacity to connect with each other. In fact, dendrites are capable of creating multiple connexions with different neurons at the same time. In order to do this they have to extend and/or shorten.
Biologists are interested to know which proteins extend or shorten the dendrites. Those proteins are in the dendrites, so we can indirectly see the dendrites because fluorophores are attached to the studied proteins.
Then, by measuring the dendrites’ length we can know if a type of protein is responsible for extending or shortening the dendrites.
There is a high interest in knowing how the extension of the dendrites works because it could greatly increase brain deceases knowledge.
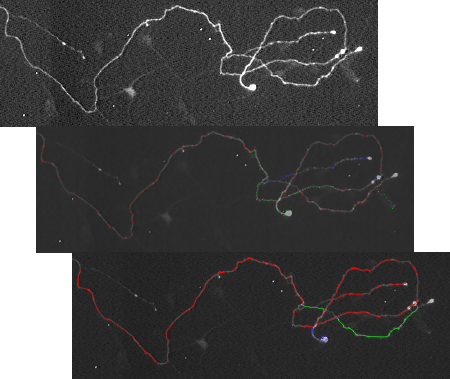
Tracking techniques
The tracking techniques are the basics of the algorithm. Starting from a given point, they iteratively search for the next point in the dendrite.
Beyond the “manual” technique, which is the one currently used by the biologists, the “box” and the “integrals” techniques are the algorithms which were implemented and tested.
Conclusion
Both tracking algorithms are working relatively well and fast. The “Integrals” technique is the most accurate but slower than the “box” technique.
However, there remain some points to improve as was said in section 4.2. The main point to improve in the code is the problem with the neuron’s core, which increase artificially the dendrite total length.
To reduce the false detections, the pre-processing should be improved to increase the difference between the background pixel value and the neuron pixel value.
This algorithm cannot be used now by biologists since the parameters are too long and to complicated to set. The pre-processing and image parameters analyse should be automated, so the only parameter would be a starting point.
In this case biologists would only have to suppress the false detections, which is done very quickly when you know which length you are looking for.
|