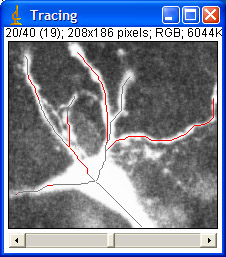
 A team of the Swiss institute for experimental cancer research (ISREC) search to quantify the synaptic activity that are able to influence the morphological development of neurons. For this, the researchers have developed a experimental model in vitro and perform some morphological studies based on confocal images. A team of the Swiss institute for experimental cancer research (ISREC) search to quantify the synaptic activity that are able to influence the morphological development of neurons. For this, the researchers have developed a experimental model in vitro and perform some morphological studies based on confocal images.
The easiest way to observe differences between treated and non- treated neurons is to:
- measure the dendrites length
- to detect the dendrite-bifurcation
- measure the angle between the parting branches
This semester project has as goal to compute and evaluate all of these information. At present, all this information are extracted manually in a tedious and time consuming fashion. The goal is to automate the extraction of information and to write a java plugin for ImageJ, a image handling software, which is able to measure the length of the axon and the dendrites, to detect dendrite-separations and to measure the angel between the branches. Finally all the information is put into an output file.
|