| Saskia Delpretti | Semester project |
| Section Sciences de la Vie, EPFL | June 2007 |
Noise distorts fluorescence digital images, altering the extraction of biological information. Once familiarized with the different noise contributions in fluorescence bioimaging and their related statistical distribution models, the goal of this project was to adapt an already existing monovariate wavelet-based denoising java code for multivariate wavelet-based denoising of fluorescence image sequences, either 3D or 2D timelapse image sequences. The multivariate wavelet-based denoising algorithm takes profit of the fact that different adjacent frames in an image sequence contain strong common information. It maximizes peak signal-to-noise ratio (PSNR) by minimizing SteinŐs unbiased risk estimate (SURE) of the mean square error between the clean and the denoised image sequence. We built an ImageJ plugin with an easy to use interface for multivariate denoising. We first validated the implemented denoising algorithm on real unnoisy biological sequences that we had corrupted with noise and proved that better denoising performances are reached with multivariate denoising compared with monovariate denoising. We then tested the implemented denoising algorithm on real noisy biological sequences and showed that the extraction of biological information can be facilitated with multivariate denoising.
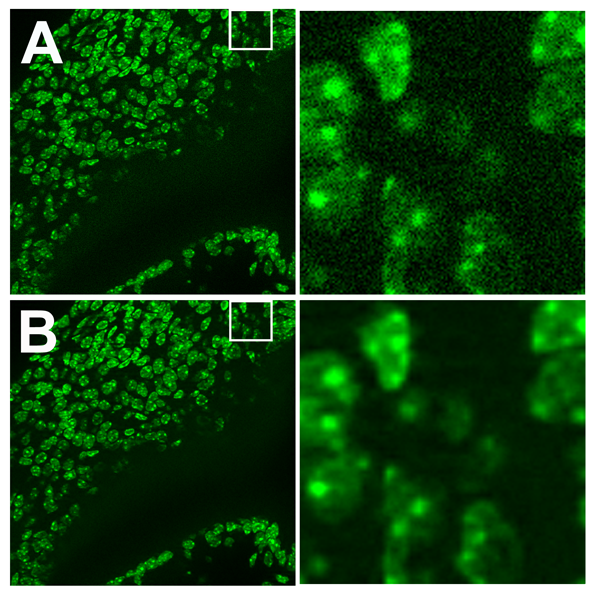
Figure 1: (A) Real noisy confocal 3D image sequence. (B) Idem A denoised with multivariate denoising and a Poisson assumption on the noise model.
Figure 2: Interface for the multiframe denoising plugin. (Standard conditions)