| EPFL STI-IMT-LIB |
| BM 4.135, Station 17 |
| CH-1015 Lausanne |
| Switzerland |
| +41 21 693 51 89 (EPFL) |
| +41 21 691 54 25 (Ecublens) |
| daniel.sage@epfl.ch |
| http://bigwww.epfl.ch/sage/ |
A software package for single particles tracking over noisy image sequence
Abstract: StopTracker is a robust and fast computational procedure for tracking fluorescent markers in time-lapse microscopy. The algorithm is optimized for finding the time-trajectory of single particles in very noisy dynamic (2D or 3D) image sequences. It proceeds in three steps. First, the images are aligned to compensate for the movement of the biological structure under investigation. Second, the particle's signature is enhanced by applying a Mexican hat filter, which we show to be the optimal detector of a Gaussian-like spot in presence of noise. Finally, the optimal trajectory of the particle is extracted by applying a dynamic programming optimization procedure.
Free Software: SpotTracker is a set a two Java plug-ins (Version 2D and Version 3D) for the public-domain ImageJ software. To run the plugin you should first download a version of ImageJ ; it only takes a couple of minutes to install ImageJ and it is available on all platforms. You'll be free to use this software for research purposes, but you should not redistribute it without our consent. In addition, we expect you to include a citation or acknowledgment whenever you present or publish results that are based on it.
Reference: D. Sage, F. Hediger, S.M. Gasser, M. Unser, "Automatic Tracking of Particles in Dynamic Fluorescence Microscopy," Proceedings of the Third International Symposium on Image and Signal Processing and Analysis (ISPA'03), Rome, Italy, September 18-20, 2003, pp. I.582-I.586.
Application: SpotTracker is used to track the movement of chromosomal loci within nuclei of budding yeast cells. The data are provided by the research group of Prof. Susan Gasser with the Departement of Molecular Biology, Unversity of Geneva.
| 2D Version | 3D Version | |||||||
Download: SpotTracker2D.zip |
Download: SpotTracker3D.zip |
|||||||
| Release: 13.10.2004 | Release: 15.04.2005 | |||||||
Data sample: Sequence2D.zip |
Data sample: Sequence3D.zip |
|||||||
| 200 frames | 1800 frames (300 times, 6 z-slices) | |||||||
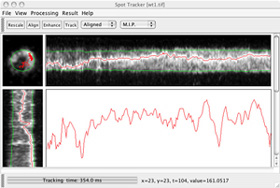 |
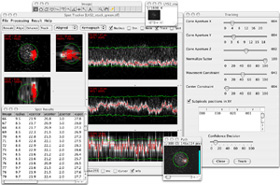 |
|||||||
| Click to enlarge the screenshot | Click to enlarge the screenshot | |||||||
Presentation: PowerPoint Slides