These synthetic data consist of six parallel hollow bars. The images have been blurred using a theoretical microscopic PSF, and corrupted by Gaussian noise and Poisson noise with several signal to noise ratios (SNR = 15, 30 dB). The knowledge of the ground-truth allows one to quantatively compare the deconvolution tools.
| Dataset | Series of 2D images | Single z-stack (TIF) |
| Uncorrupted stack (reference) | Bars.zip | Bars-stack.zip |
| Blurred and additive (low) noisy stack | Bars-G10-P15.zip | Bars-G10-P15-stack.zip |
| Blurred and additive (high) noisy stack | Bars-G10-P30.zip | Bars-G10-P30-stack.zip |
| PSF | PSF-Bars.zip | PSF-Bars-stack.zip |
| Statistics | Mininum | Maximum | Mean μ | Standard deviation |
|---|---|---|---|---|
| Size | Image size: 256x256 pixels, 128 z-slices, 1 channel, TIFF, 16-bits | |||
| Uncorrupted | 0 | 65635 | 205.59 | 3664.85 |
| G10-P15 | 0 | 65635 | 3527.12 | 4294.82 |
| G10-P30 | 0 | 65635 | 5817.16 | 7514.97 |
| Feature | Value |
|---|---|
| Type | Widefield |
| Refractive index | ni: 1.518 |
| Numerical aperture | NA: 1.4 |
| Spherical aberration | W040: 0 |
| Wavelength | lamda: 500 nm |
| Spatial resolution | deltar: 100 nm |
| Axial resolution | deltaz: 200 nm |
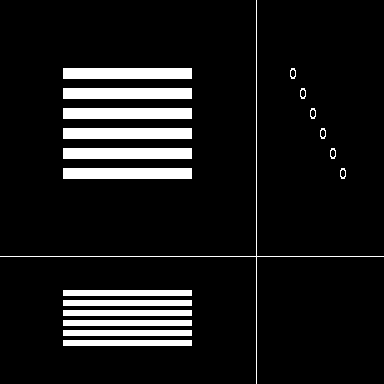 |
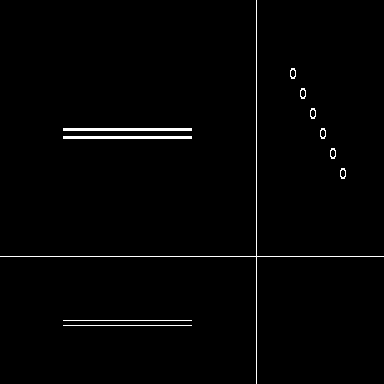 |
| Maximum intensity projection of the reference | 3 Orthogonal views of the reference |
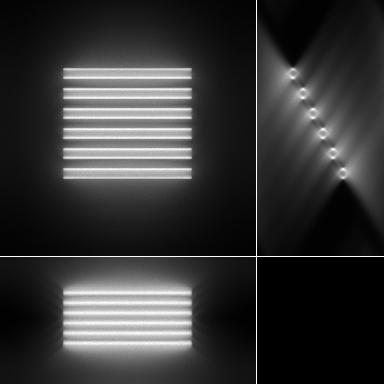 |
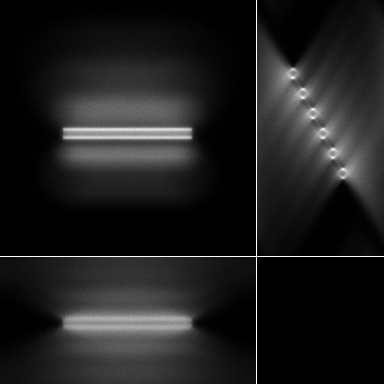 |
| Maximum intensity projection of the corrupted data | 3 Orthogonal views of the corrupted data |
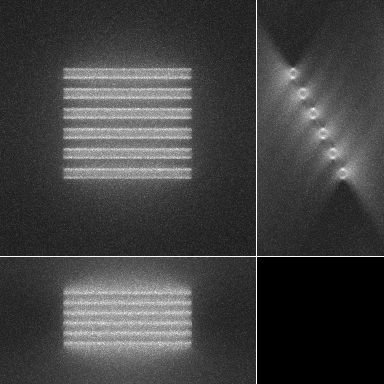 |
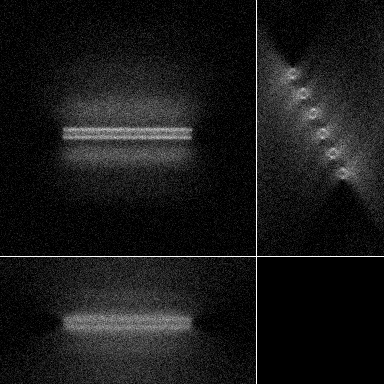 |
| Maximum intensity projection of the corrupted data | 3 Orthogonal views of the corrupted data |
 |
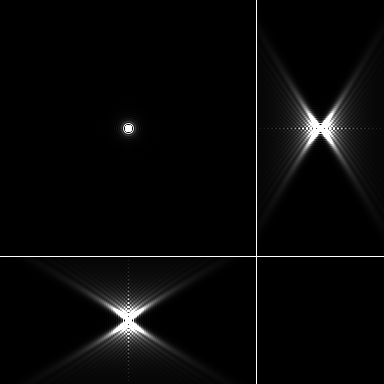 |
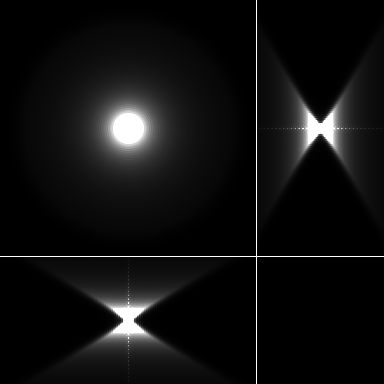
| Maximum intensity projection of the PSF | 3 Orthogonal views of the PSF |