DeconvolutionLab2 is freely accessible and open-source for 3D deconvolution microscopy; it can be linked to well-known imaging software platforms, ImageJ, Fiji, ICY, Matlab, and it runs as a stand-alone application.
The backbone of our software architecture is a library that contains the number-crunching elements of the deconvolution task. It includes the tool for a complete validation pipeline. Inquisitive minds inclined to peruse the code will find it fosters the understanding of deconvolution.
At this stage, DeconvolutionLab2 includes a friendly user interface to run the following algortihms: Regularized Inverse Filter, Tikhonov Inverse Filter, Naive Inverse Filter, Richardson-Lucy, Richardson-Lucy Total Variation, Landweber (Linear Least Squares), Non-negative Least Squares, Bounded-Variable Least Squares, Van Cittert, Tikhonov-Miller, Iterative Constraint Tikhonov-Miller, FISTA, ISTA.
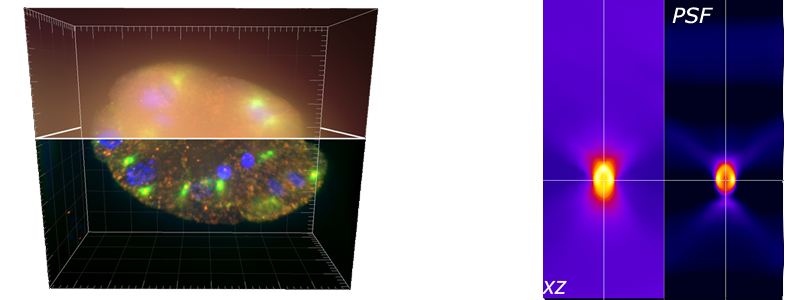
|
D. Sage, L. Donati, F. Soulez, D. Fortun, G. Schmit, A. Seitz, R. Guiet, C. Vonesch, M. Unser DeconvolutionLab2 : An Open-Source Software for Deconvolution Microscopy Methods-Image Processing for Biologists, vol. 115, 2017. |
| Download | ||
|
Version 2.1.2 (27.06.2018) Do not unzip this downloaded file |
||
| Installation | ||
ImageJ |
Put the file DeconvolutionLab_2.jar in the plugins folder and restart ImageJ or Fiji. Check the menu Plugins » DeconvolutionLab2.
Example of ImageJ macro
image = " -image synthetic Cube 10.0 1.0 size 200 100 100"
psf = " -psf synthetic Double-Helix 3.0 30.0 10.0 size 200 100 100"
algorithm = " -algorithm RIF 0.1000 -out mip MI1 -path home"
run("DeconvolutionLab2 Launch", image + psf + algorithm + parameters) |
|
ImageJ2 |
||
Fiji |
||
Matlab |
Put the DeconvolutionLab_2 into the java folder of Matlab Example of Matlab commands javaaddpath([matlabroot filesep 'java' filesep 'DeconvolutionLab_2.jar']) result = DL2.RIF(rand(40, 40, 30), rand(30, 30, 30), 0.125, '-out mip rand'); |
|
Stand-alone application |
Require a Java JDK 1.8 properly installed in the machine. Double-click on the DeconvolutionLab_2.jar file to the start the user interface of DeconvolutionLab2.
|
|
Java code |
Use the DeconvolutionLab2 as a Java library Snippet of Java code
RealSignal r = Lab.getImage("fundus.tif");
RealSignal h = new DirectionalMotionBlur(2, 20, 20).generate(r.nx, r.ny, r.nz);
Simulation sim = new Simulation(0, 1, 0);
RealSignal y = sim.run(r, h);
TikhonovRegularizedInverseFilter trif = new TikhonovRegularizedInverseFilter(0.001);
RealSignal x = trif.run(y, h);
Lab.show(x, "TRIF " + i);
|
|
Icy |
Not yet ready for the centralized distribution on the Icy website | |
Command line interface |
|
|
| Source code, documentation, previous versions | ||
| Source code on GitHub | git clone https://github.com/Biomedical-Imaging-Group/DeconvolutionLab2 | |
| History | Status of the development | |
| Library API | Java doc | |
| Version 2.0.0 | DeconvolutionLab_2-2.0.0.jar | |
| FFT Libraries | ||
| JTransforms | Visit the page of JTransforms JTransforms is already included in Fiji and Icy |
Get a JTransforms.jar file and put it in the same directory than DeconvolutionLab_2.jar |
| FFTW Version 2 |
FFTW.zip It is include the FFTW2 dynamic libraries for Mac OSX, and Windows 32-bits and 64-bits machines, and Linux 32-bits and 64-bits machines. |
Download the FFTW.zip folder, unzip it and put it in the same directory than DeconvolutionLab_2.jar |
▶ Image
▶ PSF
▶ Algorithm
▶ Path
|
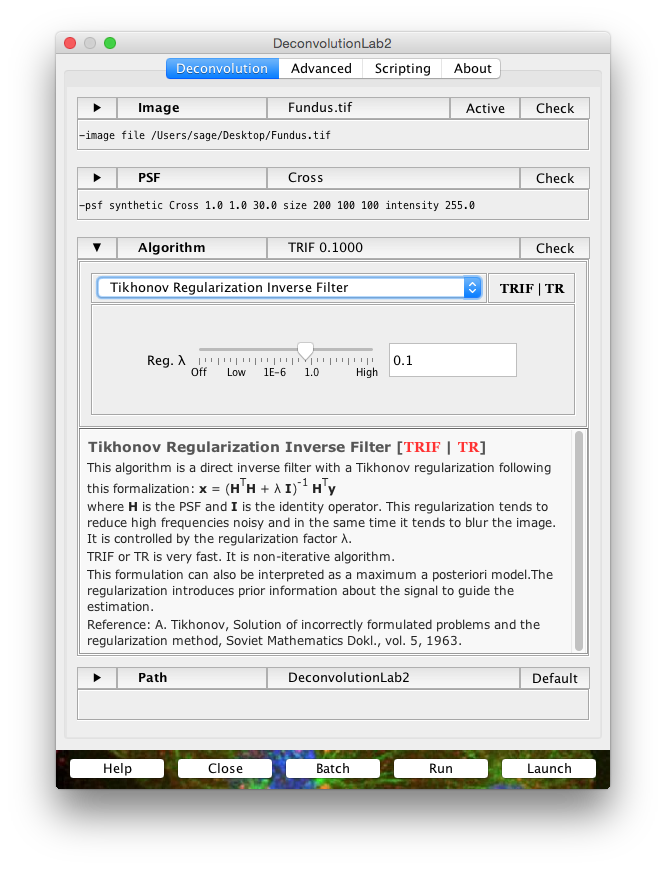
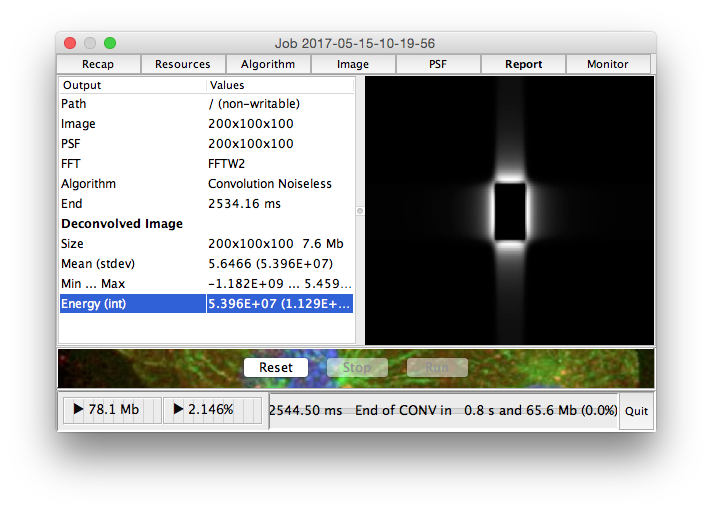

|
Run |
Run the deconvolution task in headless mode |
Launch |
Start the deconvolution dialog box, then Run the deconvolution task |
Batch |
The current deconvolution command is added in the Batch table (see Advanced tab) |
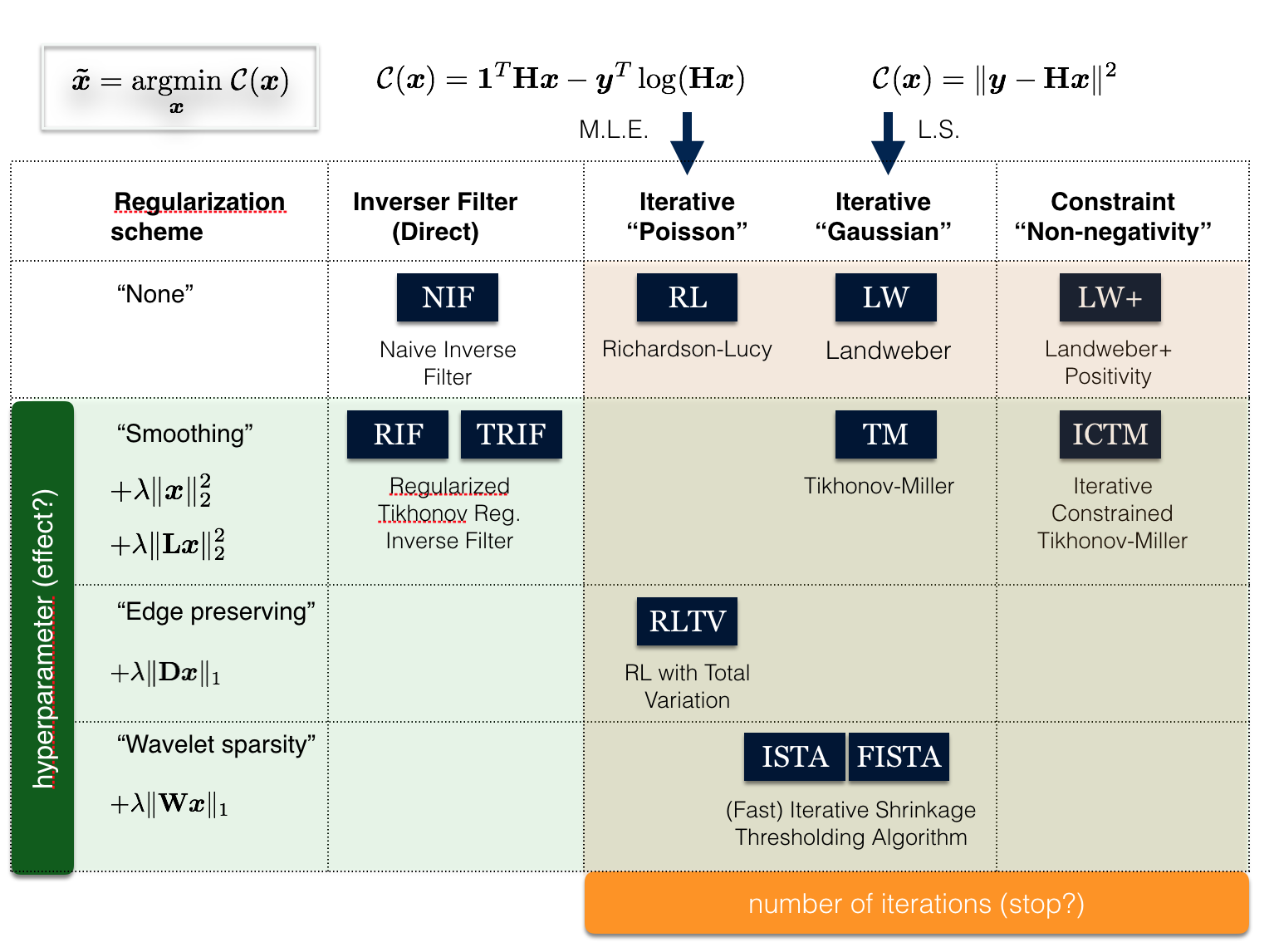
| Algorithms | Shortname | Iterative | Step Controllable | Regularization | Wavelets |
| Deconvolution | |||||
| Regularized Inverse Filter Laplacian Regularized Inverse Filter |
RIF LRIF |
Direct | No | Yes | |
| Tikhonov Regularized Inverse Filter | TRIF | Direct | No | Yes | |
| Naive Inverse Filter Inverse Filter |
NIF IF |
Direct | No | Yes | |
| Richardson-Lucy | RL | Iterative | No | No | |
| Richardson-Lucy Total Variation | RLTV | Iterative | No | Yes | |
| Landweber Linear Least Squares |
LW LLS |
Iterative | Yes | No | |
| Non-negative Least Squares Landweber+Positivity |
NNLS LW+ |
Iterative | Yes | No | |
| Bounded-Variable Least Squares Spark-Parker |
BVLS SP |
Iterative | Yes | No | |
| Van Cittert | VC | Iterative | Yes | No | |
| Tikhonov-Miller | TM | Iterative | Yes | Yes | |
| Iterative Constraint Tikhonov-Miller | ICTM | Iterative | Yes | Yes | |
| FISTA | FISTA | Iterative | No | Yes | Haar, Spline |
| ISTA | ISTA | Iterative | No | Yes | Haar, Spline |
| Simulation | |||||
| Simulation | SIM | Direct | Convolution with a PSF and corruption with Gaussian and Poisson noise | ||
| Convolution | CONV | Direct | Convolution with a PSF (no additive noise) | ||
| Identity | I | Direct | Copy the input into the output | ||
| Non-stabilized Division | DIV | Direct | Division by a PSF in the Fourier domain | ||
The command line of DeconvolutionLab2 consists of a series of arguments that allows a full control of the processing. The command line is written in a single line, space is mostly used as separator. The general format of the argument is:
-keyword [option] parameters
The list of keywords and the options presenting in the following table. The sign | indicate a OR). The default value are written in bold
| Keyword | Default | Options | Description |
| -image file | Mandatory | Path to a single file (z-stack) usually a TIF or STK file |
Source of images. The 3D input data should be a z-stack of images. |
| -image directory | Path to a directory containing 2D images [pattern |
||
| -image synthetic | Name and parameters of the shape [intensity, size, center]> |
||
| -image platform | Name of the image of the platform (ImageJ or Icy) | ||
| -psf file | Mandatory | Path to a single file (z-stack) used as PSF usually a TIF or STK file |
Source of PSF. The 3D PSF data should be a z-stack of images. |
| -psf directory | Path to a directory containing 2D images [pattern] |
||
| -psf synthetic | Name and parameters of the shape [intensity, size, center] |
||
| -psf platform | Name of the image of the platform (ImageJ or Icy) | ||
| -algorithm | Mandatory |
RIF | TRIF | NIF | LW | NNLS | BVLS | RL | RLTV | TM | ICTM | ISTA | FISTA | VC | I | CONV | SIM | DIV
Synonym of the acronym: RIF = LRIF, NIF = IF, LW = LLS, NNLS = LW+, BVLS = SP, I = ID |
Name and parameter of the algorithm |
| -path | current | current | path | Working directory |
| Output (several instances of out are possible) | |||
| -out stack | intact float |
Name of the output: Note that this name is used as title of the window image and as the filename for the storage Option for dynamic: intact | rescaled | normalized | clipped Option for type: byte | short | float Mode: By default the output is shown and saved. nosave | noshow |
Output as a stack of images (TIF) |
| -out series | intact float | Output as series of 2D images (slices, XY) | |
| -out mip | intact float | Output as a maximum-intensity projection | |
| -out ortho | intact float | Output as a 3 orthogonal views centered around the keypoint | |
| -out planar | intact float | Outputs as a 2D side-to-side image of all the z-slices | |
| Controller | |||
| -monitor | console table | console | table | no | Selection of the monitoring output |
| -verbose | log | log | quiet | prolix | mute | Message monitoring |
| -stats | show | show | save | no | Statistics |
| -constraint | no | no | nonnegativity | clipped | Spatial constraint on the signal |
| -residu | no | no | value | Stops when the minimal residu is reached |
| -time | no | no | value | Limitation of running time |
| -reference | no | no | filename | Assess the current deconvolved image with the reference image |
| Preprocessing | |||
| -pad | NO NO 0 0 | NO | X2 | X23 | X235 | E2 | Lateral and axial padding and extension scheme |
| -apo | NO NO | UNIFORM | NO | HAMMING | HANN | COSINE | TUKEY | WELCH | Lateral and axial apodization window function |
| -norm | 1 | no | value | Normalization factor for the PSF |
| Resources | |||
| -fft | fastest | academic | jtransforms | fftw2 | Indicates the FFT library |
| -epsilon | 1E-6 | value | Machine Epsilon |
Example
-image synthetic Cube 10.0 1.0 size 200 100 100 intensity 255.0 -psf synthetic Double-Helix 3.0 30.0 10.0 size 200 100 100 intensity 255.0 -algorithm RIF 0.1000 -out mip MI1 -norm 10.0 -path home
Paul Kefer and Pierre-Alexandre Vidi, Dept of Cancer Biology at the Wake Forest University Baptist Medical Center, have written a ImageJ macro to deconvolve a series of frames by the algorithm RIF of DeconvolutionLab2.
Download the Image Macro: time_lapse_deconvolvution_rif.ijm| Material | Download | Size | Description |
| Slide in PDF (without the animation/video) | 3D-Deconvolution-Microscopy.pdf | 4.6 Mb | Course given in Neubias 2020, February 2017 |
| Restoration | logo.zip | 0.6 Mb | 2D simulation, influence of the PSF shape to restore the original image |
| Naive Inverse Filter | naive-deconvolution.zip | 3.3 Mb | 2D simulation of the inverse filter |
| Simulation to check the resolution | test-resolution.ijm | 0 Mb | Macro to generate simulated data and simulated PSF |
| Simulation to check the spectral effect | spectral-analysis.zip | 0.7 Mb | 2D simulation of fine structures |
| Hollow bars | bars.zip | 28 Mb | 3D reference, 3D corruputed data and 3D PSF |
| Synthetic microtubules | microtubules-challenge.zip | 63 Mb | 3D data and 3D theoretical PSF of a realistic specimen |
| Drosophila (crop) | drosophila-crop.zip | 19 Mb | Small 3D data and 3D theoritical PSF |
You'll be free to use this software for research purposes, but you must not transmit and distribute it without our consent. In addition, you undertake to include a citation whenever you present or publish results that are based on it. EPFL makes no warranties of any kind on this software and shall in no event be liable for damages of any kind in connection with the use and exploitation of this technology.