C. elegans embryo
This real dataset is composed of three stacks of images of a C. Elegans embryo.
The deconvolution effects can be evaluated on different kinds of structures: extended objects (the chromosomes in the nuclei),
filaments (the microtubules), and point-wise spots (a protein stained with CY3).
Data
| Statistics |
Mininum |
Maximum |
Mean μ |
Std deviation |
| CY3 Channel |
215 |
2842 |
992.34 |
595.23 |
| FITC Channel |
209 |
2929 |
657.66 |
327.59 |
| DAPI Channel |
205 |
2687 |
585.75 |
290.13 |
PSF
| PSF Features
| | Type | Widefield |
| Refractive index | ni: 1.518 |
| Numerical aperture | NA: 1.4 |
| Spherical aberration | W040: 0 |
| Wavelength | DAPI:477nm; FITC:542nm; CY3:654nm |
| Spatial resolution | deltar: 64.5 nm |
| Axial resolution | deltaz: 160 nm |
|
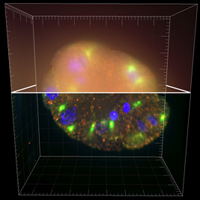
Image size: 672x712 pixels
Number of z slices: 104 slices
Number of channels: 3
Dynamic: 16 bits
Image format: TIFF
|
Microscope Acquisition: Olympus CellR
- Objective: UPlanSApo 100X/1.4 oil(n.i. 1.518); image pixel size (binning 1X1): 64.5 nm
- Laser intensity: 100% - Camera pixel size: 6450 nm - Gain: 0
- z step: 200 nm (experiment 10, 1344x1024x111; crop 672x712x104)
- Channel 1: DAPI, lamda excitation: 377/50 nm, lamda emission: 447/60 nm; Channel 2: FITC, lamda excitation: 485/20, lamda emission: 531/22; Channel 3: CY3,lamda excitation: 560/25,lamda emission: 634/40
Orthogonal preview
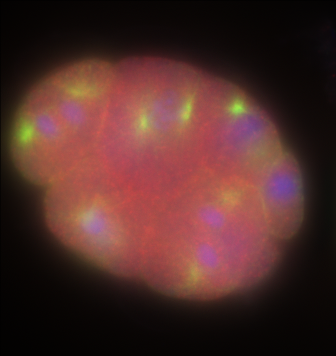
XY section |

ZY section |
XZ section |


