GlobalBioIm
Global integrative framework for Computational Bio-Imaging
Principal Investigator: Prof. Michael Unser
Participants: Shayan Aziznejad, Thomas Debarre, Laurene Donati, Julien Fageot, Harshit Gupta, Kyong Jin, Thanh-An Pham, Emmanuel Soubies, Ferreol Soulez, Luc Zeng
Period: October 2016-October 2021.
Abstract
A powerful strategy for increasing the quality and resolution of medical and biological images is to acquire larger quantities of data (Fourier samples for MRI, projections for X-ray imaging) and to jointly reconstruct the complete signal by correctly reallocating the measurements in 3D space/time and integrating all the information available. The underlying image sequence is reconstructed globally as the result of a very large-scale optimization that exploits the redundancy of the signal (sparsity, spatio-temporal correlation) to improve the solution. Due to recent advances in the field, we are arguing that such a "bigger data" integration is now within reach and that our team is ideally qualified to lead the way. A successful outcome will profoundly impact the design of future bioimaging systems.
We are proposing a unifying framework for the development of such next-generation reconstruction algorithms with a clear separation between the physical (forward model) and signal-related (regularization, incorporation of prior constraints) aspects of the problem. The pillars of our formulation are: an operator algebra with a corresponding set of fast linear solvers; an advanced statistical framework for the principled derivation of reconstruction methods; and learning schemes for parameter optimization and self-tuning. These core technologies will be incorporated into a modular software library featuring the key components for the implementation and testing of iterative reconstruction algorithms. We shall apply our framework to improve upon the state of the art in the following modalities:
- phase-contrast X-ray tomography in full 3D;
- structured illumination microscopy;
- single-particle analysis in cryo-electron tomography;
- a novel multipose fluorescence microscopy;
- real-time MRI, and
- a new multimodal digital microscope.
In all instances, we shall work in close collaboration with the imaging scientists who are in charge of the instrumentation.

GlobalBioIm Library
A unifying Matlab library for imaging inverse problem
When being confronted with a new inverse problem, the common experience is that one has to reimplement (if not reinvent) the wheel (=forward model + optimization algorithm), which is very time consuming and also acts as a deterrent for engaging in new developments. This Matlab library GlobalBioIm aims at simplifying this process by decomposing the workflow onto smaller modules, including many reusable ones since several aspects such as regularization and the injection of prior knowledge are rather generic. It also capitalizes on the strong commonalities between the various image formation models that can be exploited to obtain fast, streamlined implementations.
Open Access to the GlobalBioIm LibraryReferences
[1] M. Unser, E. Soubies, F. Soulez, M. McCann, L. Donati, GlobalBioIm: A Unifying Computational Framework for Solving Inverse Problems Proceedings of the OSA Imaging and Applied Optics Congress on Computational Optical Sensing and Imaging (COSI'17), San Francisco CA, USA, 2017.
[2] E. Soubies, F. Soulez, M.T. McCann, T.-a. Pham, L. Donati, T. Debarre, D. Sage, M. Unser, Pocket Guide to Solve Inverse Problems with GlobalBioIm, Inverse Problems, vol. 35, no. 10, pp. 1-20, October 2019.
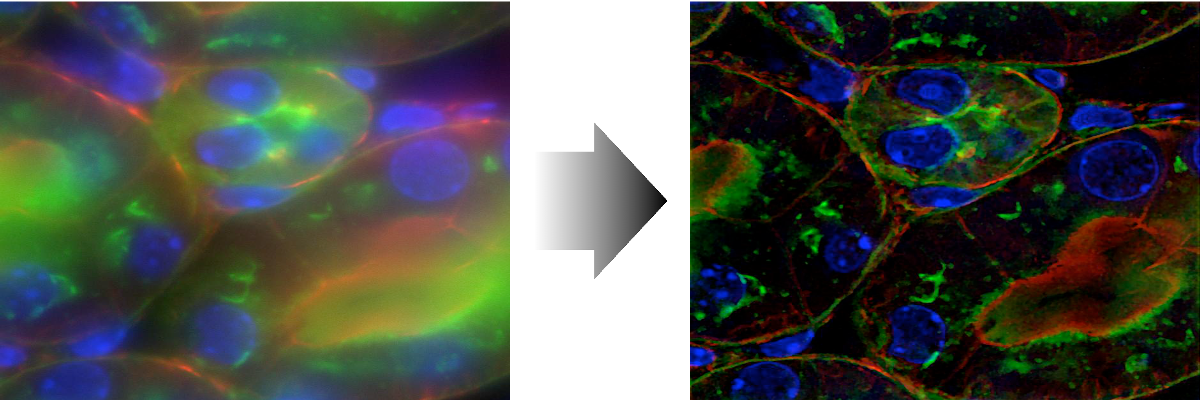
3D deconvolution microscopy
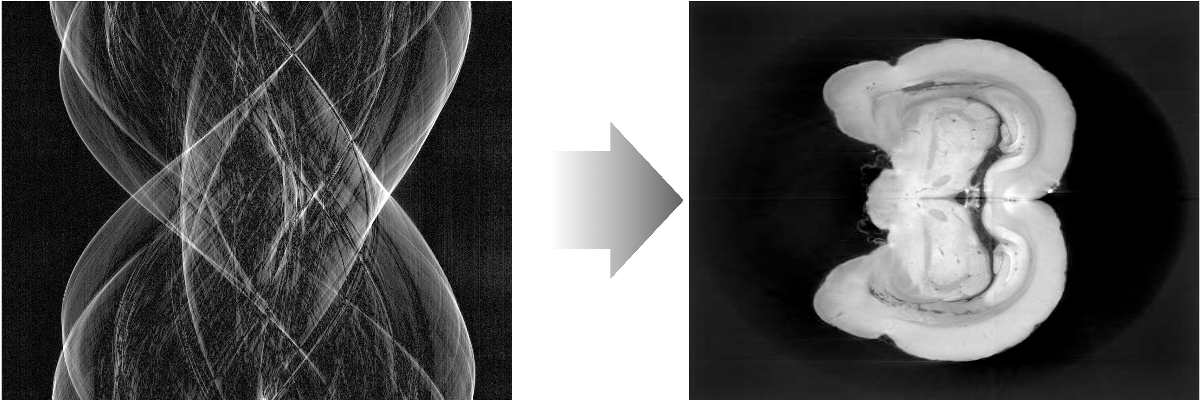
Tomography reconstruction