Identification of Point Spread Function (PSF) aberrations using genetic algorithms
Spring 2011
Master Semester Project
Master Diploma
Project: 00206
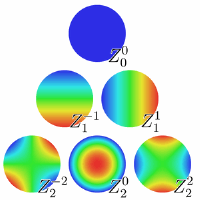
In this project, our motivation is to develop an algorithm and some associated software that helps identifying possible causes of optical aberrations that can exist in certain microscopes (e.g. widefield or confocal), due to non-ideal optical elements and/or bad calibration.
In the context of optical microscopy, the system is characterized by its 3D response which is called the Point Spread Function (PSF). When certain defects are present in the optical system (e.g., misalignments, poor quality of certain elements, etc.), this PSF can be heavily distorted and/or asymmetric due to the associated optical aberrations. This can lead to irreversible losses in imaging quality (i.e., even after numerical corrections).
Therefore, identifying the possible sources of such distortions can help to eliminate such problems in the first place by recalibrating, correcting and/or replacing the corresponding optical elements. The identification problem amounts to finding the type of aberrations that are associated with a given (i.e., measured) intensity PSF. In order to do so, we consider a PSF forward model that is suitable for microscopy (e.g., the Richards and Wolf model [1]), in which the aberration function is parameterized by Zernicke polynomials.
The solution we want to find consists in the aberration parameters; their values must yield a PSF (computed according to the forward model) that closely matches the measured one, considering an appropriate discrepancy measure (e.g., SNR).
We shall consider a heuristic algorithm to recover the solution parameters, typically a genetic algorithm. Such an approach is innovative but required given the complexity of the problem and the number of parameters in hand. Note that, in that context, having a forward model that is computationally efficient is an important aspect. In order to gain time, we may use already-existing code or optimization toolboxes.
Throughout the project, the student might collaborate with the BioImaging and Optics platform of the Life Science faculty, in order to validate the results and/or to have some additional feedback and discussions. The code might be developed in MATLAB and/or JAVA.
Prerequisites: A good knowledge in MATLAB and/or JAVA programming is required. Ideally, having followed the course on image processing taught by M. Unser is a plus. In all cases, the student must be strongly interested to work on a project that overlaps with several fields of research, i.e., algorithmic design, optics, and signal processing.
[1] B. Richards, E. Wolf, "Electromagnetic diffraction in optical systems II. Structure of the image field in an aplanatic system"
- Supervisors
- bourquard
- Michael Unser, michael.unser@epfl.ch, 021 693 51 75, BM 4.136