Spatiotemporal Segmentation of Migrating Cells in Fluorescence Microscopy
Spring 2011
Master Semester Project
Project: 00210
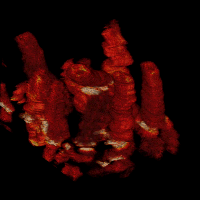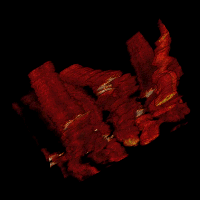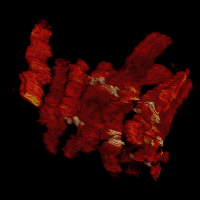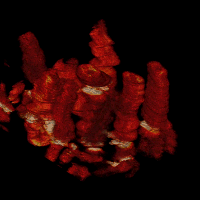
Cell dynamics is a crucial aspect of many biological processes with direct implication in human health. Changes in cellular morphology also play a critical role in the recognition of immune cells of antigens. To reliably assess the clinical potential of candidate drugs, it is usually necessary to analyze quantitatively the motion and morphology changes of cells under variety of experimental conditions. In these experiments, the temporal resolution is usually high. This allows one to display the time-lapse sequences of images as a 2D+t volumetric data. In the resulting volumes, tubes representing the migrating cells can be easily observed.
The goal of this project is to implement an ImageJ plugin to extract these spatiotemporal tubes in order to quantify the movement and morphology of the corresponding cells. To achieve that, one strategy is to use an active contour algorithm (snake) combined with the 2D+t information available.
- Supervisors
- Ricard Delgado-Gonzalo, ricard.delgado@epfl.ch, 021 693 51 43, BM 4.141
- Michael Unser, michael.unser@epfl.ch, 021 693 51 75, BM 4.136