Quantitation of asymmetric mitosis by tracking the spindle poles using fluorescence images
Spring 2011
Master Semester Project
Project: 00214
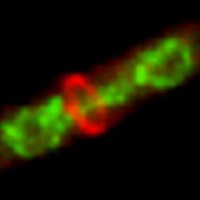
Asymmetric division is of fundamental importance during the development and maintenance of a multicellular organism. However, it is also clear that even in single-celled organisms such as yeasts, the properties of the daughter cells of a division are not identical. While the purpose and mechanism of some of these events are well-understood, for example, switching of mating type, there are other examples whose mechanism and purpose remains enigmatic. Among these are the behavior of the signaling cascade that regulates cytokinesis, which is named the SIN. The proteins are located on the poles (SPBs) of the mitotic spindle, and show an asymmetric segregation to either the old or new SPB during anaphase. The long-term goal of this project is to use analysis of time-lapse imaging of yeast cells to understand the behavior of the proteins regulating cytokinesis, and how it responds to internal and external perturbations.
The purpose of this study is to design and implement a completed image-analysis system including 1) preprocessing (denoising) 2) segmentation of the cells (active contours) 3) robust spot detector and 4) tracking phase and finally the results have to be presented in a kymograph form. All these operations can be developed as Java plugin for ImageJ.
Collaboration with Prof. Viesturs Simanis, SV-ISREC, EPFL, and Prof Ioannis Xenarios (UNIL-SIB).
- Supervisors
- Daniel Sage, daniel.sage@epfl.ch, 021 693 51 89, BM 4.135
- Michael Unser, michael.unser@epfl.ch, 021 693 51 75, BM 4.136