Modelling mycobacteria behavior in time-lapse microscopy images
Spring 2014
Master Semester Project
Master Diploma
Project: 00281
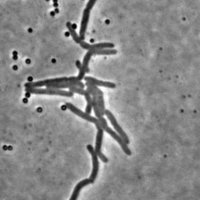
Mycobacteria are non-motile rod-shaped organisms of strong interest for tuberculosis research. As growth rate, division rate and response to external perturbations seem to vary between individuals of a same colony, there is a growing need for single cell-based analysis approaches.
The goal of this project is to propose a model for the behavior observed in time-lapse microscopy images of mycobacteria. Synthetic data will be produced using the model and compared with real-life biological images. Having access to an appropriate model would be highly valuable as it would allow creating ground truth for tracking and segmentation algorithms. It could be used as a basis for the design of automated analysis methods, and might as well be valuable for direct biological research. Synthetic images generated with such model would serve as a basis to test and compare an algorithm's performance versus manual annotation, an essential step in order to assess the efficiency of the method.
The work is very interdisciplinary as it encompasses biology, mathematics and programming aspects: the main steps of the project will be to familiarize with the literature in order to understand the physical properties of mycobacteria growth, to design a dynamic model based on this knowledge, and to implement the model in order to generate images.
- Supervisors
- Virginie Uhlmann, virginie.uhlmann@epfl.ch, 021 693 1136, BM 4.142
- Michael Unser, michael.unser@epfl.ch, 021 693 51 75, BM 4.136