Quantification of fiber-like structures in time-lapse fluorescence microscopy images
Autumn 2016
Bachelor Project
Project: 00324
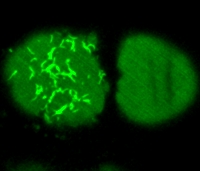
Asymmetric division is crucial for the development of multicellular organisms. The embryo of the nematode C. elegans is a well-suited model system to analyze the mechanisms of asymmetric cell division. Using spinning disc confocal microscopy, we performed live imaging of green fluorescent-tagged force generation proteins of C. elegans zygotes. This revealed that such proteins form dynamic fiber-like structures as well as foci. This student project entails designing image-analysis algorithms and applying these methods to monitor the behavior of these structures in a quantitative manner over the sequence of images. The project will be implemented in Java as a ImageJ or Icy plugin.
Collaboration with Melina Scholze, Gonczy Lab (http://gonczy-lab.epfl.ch)
- Supervisors
- Daniel Sage, daniel.sage@epfl.ch, 021 693 51 89, BM 4.135
- Michael Unser, michael.unser@epfl.ch, 021 693 51 75, BM 4.136