An off-the-grid algorithm in ImageJ for 3D single-molecule localization microscopy
Autumn 2019
Master Semester Project
Project: 00364
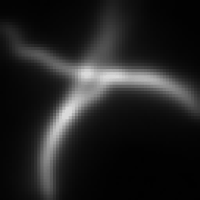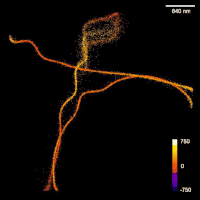
A new class of algorithms has recently emerged in the literature for the recovery of point source signals from altered and noisy measurements. These methods are able to perform the reconstruction, without requiring any discretization of the domain, by solving an infinite dimensional optimization problem. They interleave convex optimization updates (corresponding to adding a new point source) with non-convex optimization steps (corresponding to changing the intensities and positions of the point sources). Single-molecule localization microscopy is an imaging technique in fluorescence microscopy that is able to bypass the diffraction limit and reach nanoscale resolution for the imaging of sub-cellular structures in cells (e.g., microtubules). High performance numerical solvers are needed to locate precisely the positions of the fluorescent molecules. The previously mentioned class of algorithms are currently the one that obtain the state-of-the-art results in this application. The goal of this project is to implement one of these methods, called the Sliding Frank-Wolf algorithm, in Java as an ImageJ/Fiji plugin so that it can be usable by biologists. It will include a user interface and permit automatic processing of large datasets and output super-resolved images. This project is a continuation of the semester project of Amandine Evard. The code and material are available. The idea is to add new features to the algorithm and the plugin (spline interpolation of the point-spread function, tracking of molecules in time, new modalities...).
- Supervisors
- Quentin Denoyelle, quentin.denoyelle@epfl.ch, BM 4.140
- Michael Unser, michael.unser@epfl.ch, 021 693 51 75, BM 4.136
- Thanh-An Pham