Deep Learning for Angle Estimation in Cryo-EM
Autumn 2019
Master Semester Project
Project: 00365
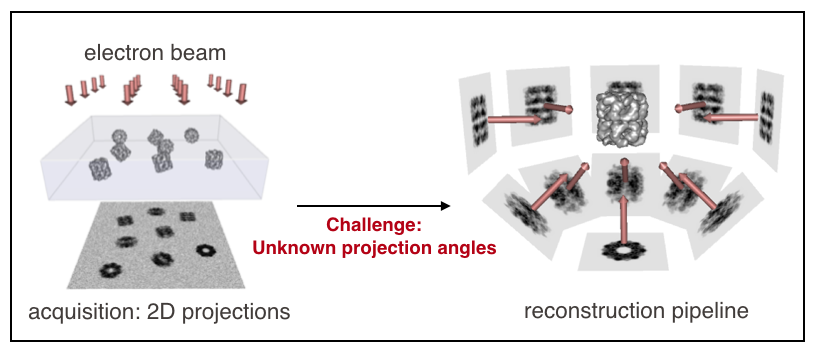
Single-particle cryo-electron microscopy (cryo-EM) is a Nobel-prized technology that aims to characterize the 3D structure of proteins at the atomic level. The electron microscope first images numerous (~100k) replicates of a protein, positioned at various orientations. Algorithms then reconstruct a high-resolution 3D structure from the acquired images.
The main challenge in cryo-EM reconstruction, compared to traditional tomographic set-ups, is that the angles at which the images were taken are unknown. Another challenge is that the images are extremely noisy and blurred. The sheer amount of images per protein (~100k), as well as the number of imaged proteins (~4k), should however enable a data-driven approach to overcome those challenges.
Project goal: Design a neural network to estimate the angular relation between images of a protein. The developed neural network will be trained and tested on simulated and real data.
Prerequisites: Experience with Python programming. Experience with (Deep) Machine Learning (with any framework) is desirable. No experience in biology is required. Experience in imaging is a plus.
- Supervisors
- Laurène Donati, laurene.donati@epfl.ch, BM 4.139
- Michael Unser, michael.unser@epfl.ch, 021 693 51 75, BM 4.136
- Michaël Defferrard (LTS2)