Characterization of myelin structure using image analysis techniques
Context
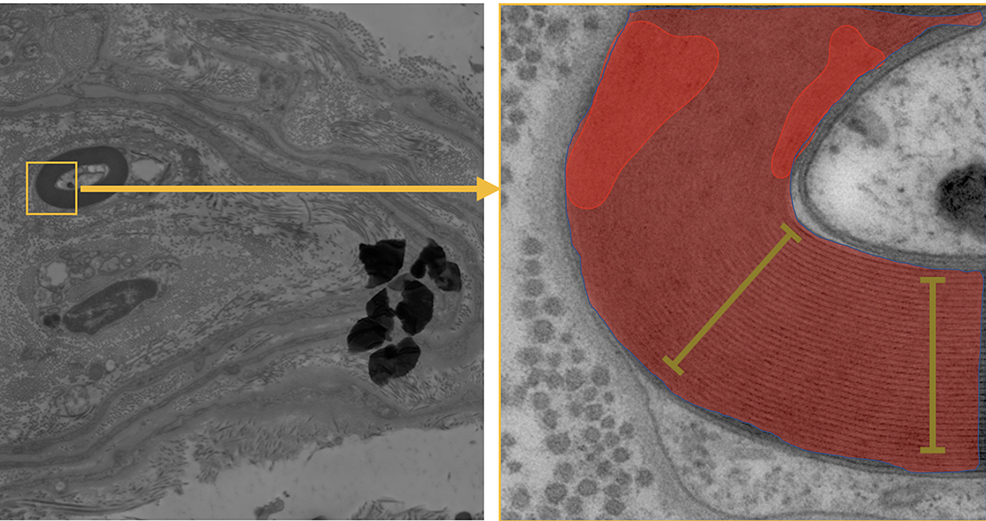
Room-temperature electron microscopy allows to study the ultrastructure of biological tissue and cells with nanometric resolution. In this project, this technique is used in particular to study the fine structure of myelinated fibers from healthy subjects and those diseased with a set of neurodegenerative diseases. In some myelinated fibers the structure is nicely preserved, while in some of them damage and swellings are visible in electron microscopy images. The goal of the project is quantify, analyze and categorize them according to their level of preservation using image analysis teachniques. This approach would be crucial to verify if myelin damage shows any correlation with the diseases.
Development
The goal of the project is to develop an image analysis workflow in Python to efficiently detect and segment all the myelinated fibers. This will enable to quantify the structure of the fibers and the damages areas in order to categorize the level of preservation. The methodology involves employing various image analysis techniques, such as denoising, edge and ridges detection, active contours, pixel classification, machine learning.
Requirement
Basic knowledge of signal or image processing
Basic skills in coding
This project is a collaboration with the EPFL Center for Imaging and the Laboratory of Biological Electron Microscopy
- Supervisors
- Daniel Sage, daniel.sage@epfl.ch
- Marta Di Fabrizio, marta.difabrizio@epfl.ch