Automated image analysis of tau fibrils in the human Alzheimer's disease brain
Reserved
Master Semester Project
Project: 00452
Context
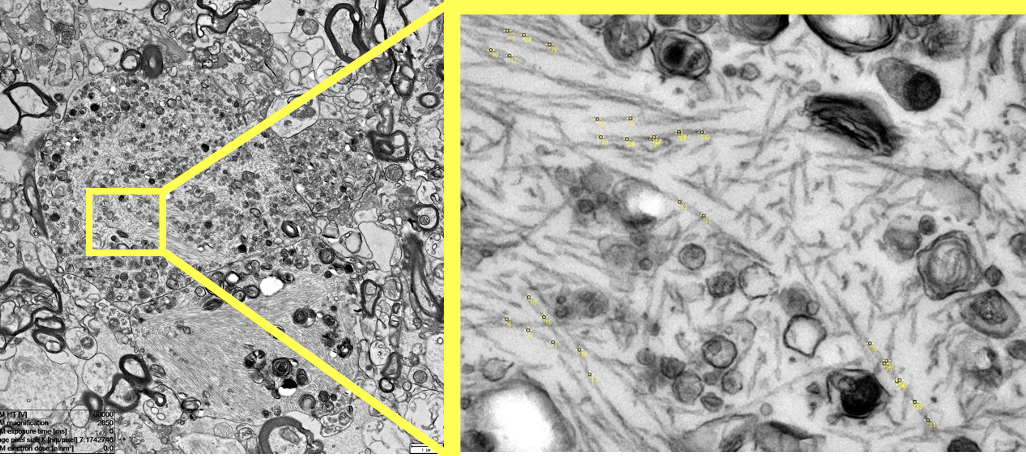
Alzheimer's disease (AD) is a devastating neurodegenerative disease affecting millions of people worldwide and costing hundreds of billions of dollars each year. To date, despite major efforts there is no cure. Two major hallmarks of AD are the aggregation of the proteins amyloid beta (Aβ) into plaques and hyperphosphorylated tau into neurofibrillary tangles (NFT). Tau fibrils in such tangles can adopt different structures called conformer. A key feature of such conformers is the twist of these helical tau fibrils. In this project I investigate the different stages of tau tangles on an ultrastructural level directly in the human brain using correlative light and electron microscopy. The goal of this project is to detect tau fibrils within the tau tangles and measure their twist. It will be interesting to see if the twist is stage dependent and if tau fibrils of the same type cluster together. This could inform us about the aggregation process of tau tangles directly the human brain.
Development
The goal of the project is to develop an image analysis workflow in Python to efficiently detect and segment tau fibrils. This will allow us to measure the twist of tau fibrils and put it in a cellular/tissue context. This project involves employing various image analysis techniques, such as denoising, line detection, active contours, pixel classification, machine learning. Requirement
Basic knowledge of signal or image processing Basic skills in coding
This project is a collaboration with the EPFL Center for Imaging and the Laboratory of Biological Electron Microscopy
Alzheimer's disease (AD) is a devastating neurodegenerative disease affecting millions of people worldwide and costing hundreds of billions of dollars each year. To date, despite major efforts there is no cure. Two major hallmarks of AD are the aggregation of the proteins amyloid beta (Aβ) into plaques and hyperphosphorylated tau into neurofibrillary tangles (NFT). Tau fibrils in such tangles can adopt different structures called conformer. A key feature of such conformers is the twist of these helical tau fibrils. In this project I investigate the different stages of tau tangles on an ultrastructural level directly in the human brain using correlative light and electron microscopy. The goal of this project is to detect tau fibrils within the tau tangles and measure their twist. It will be interesting to see if the twist is stage dependent and if tau fibrils of the same type cluster together. This could inform us about the aggregation process of tau tangles directly the human brain.
Development
The goal of the project is to develop an image analysis workflow in Python to efficiently detect and segment tau fibrils. This will allow us to measure the twist of tau fibrils and put it in a cellular/tissue context. This project involves employing various image analysis techniques, such as denoising, line detection, active contours, pixel classification, machine learning. Requirement
Basic knowledge of signal or image processing Basic skills in coding
This project is a collaboration with the EPFL Center for Imaging and the Laboratory of Biological Electron Microscopy
- Supervisors
- Daniel Sage, daniel.sage@epfl.ch
- Daniel Stähli, daniel.stahli@epfl.ch