Analysis of dendritic spine changes using deep-learning encoding in a latent space
Reserved
Master Semester Project
Master Diploma
Project: 00453
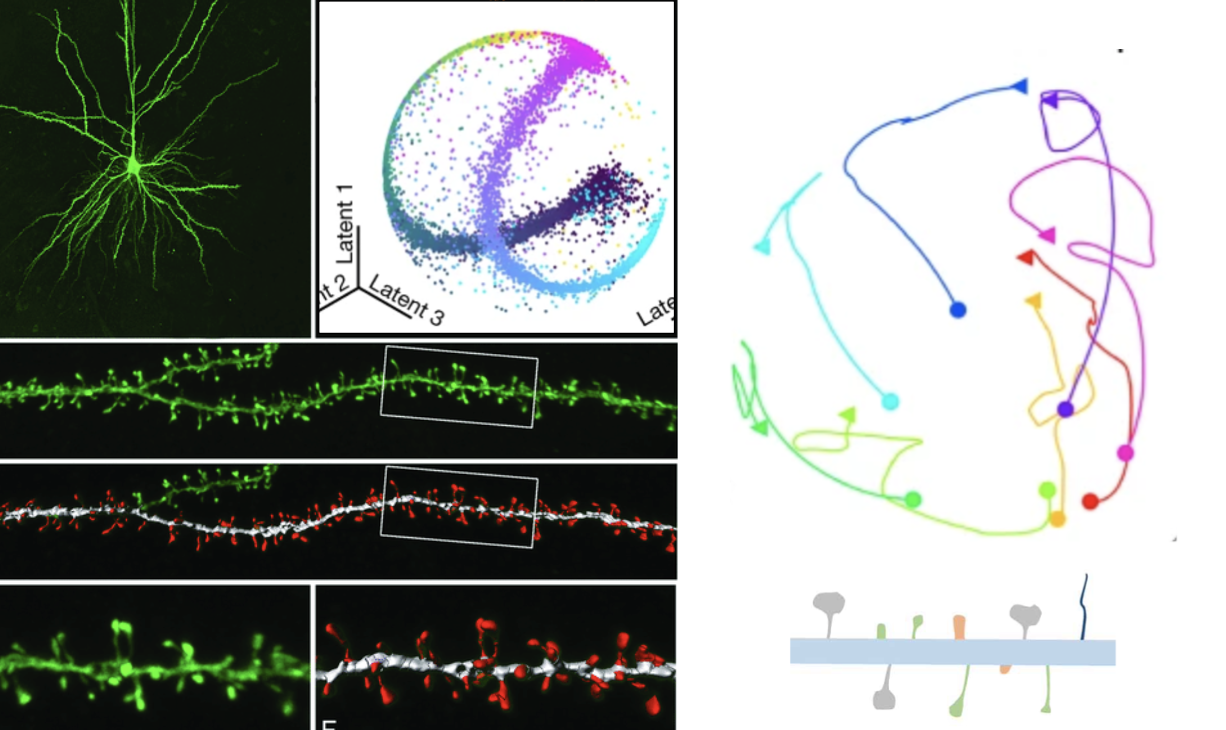
The dynamic changes in dendritic spine morphology play a crucial role in enhancing the understanding of neural plasticity. These changes serve as indicators of the cellular processes underlying learning and memory, and they are symptomatic in various neurodegenerative disorders. The new time-lapse microscopy techniques now allow the acquisition of high-resolution 3D images of spines. Moreover, advanced image analysis methods have facilitated the segmentation of dendritic spines in 3D along the dendrites.
This project aims to develop a system able to accurately reconstructing the evolution of dendritic spines. By using a large dataset of spine shapes recorded at different time points, the system will ultimately enable the classification of single spine behaviors. Here we propose to use a deep learning approach to guess the intricate evolution of dendritic spines. We will encode the spine shapes into a low-dimensional latent space to capture the most important temporal features required to represent the input data. The project includes an exploratory phase to determine the optimal dimensions of the latent space and the types of trajectories that best explain the spine's morphometry. This innovative methodology has the potential to shed light on the dynamics of neural plasticity.
Coding: Python / Pytorch
- Supervisors
- Aleix Boquet I Pujadas, aleix.boquetipujadas@epfl.ch
- Daniel Sage, daniel.sage@epfl.ch