From PDE to Cell Imaging: A Multiresolution Framework for Fourier Ptychography
Master Semester Project
Master Diploma
Project: 00454
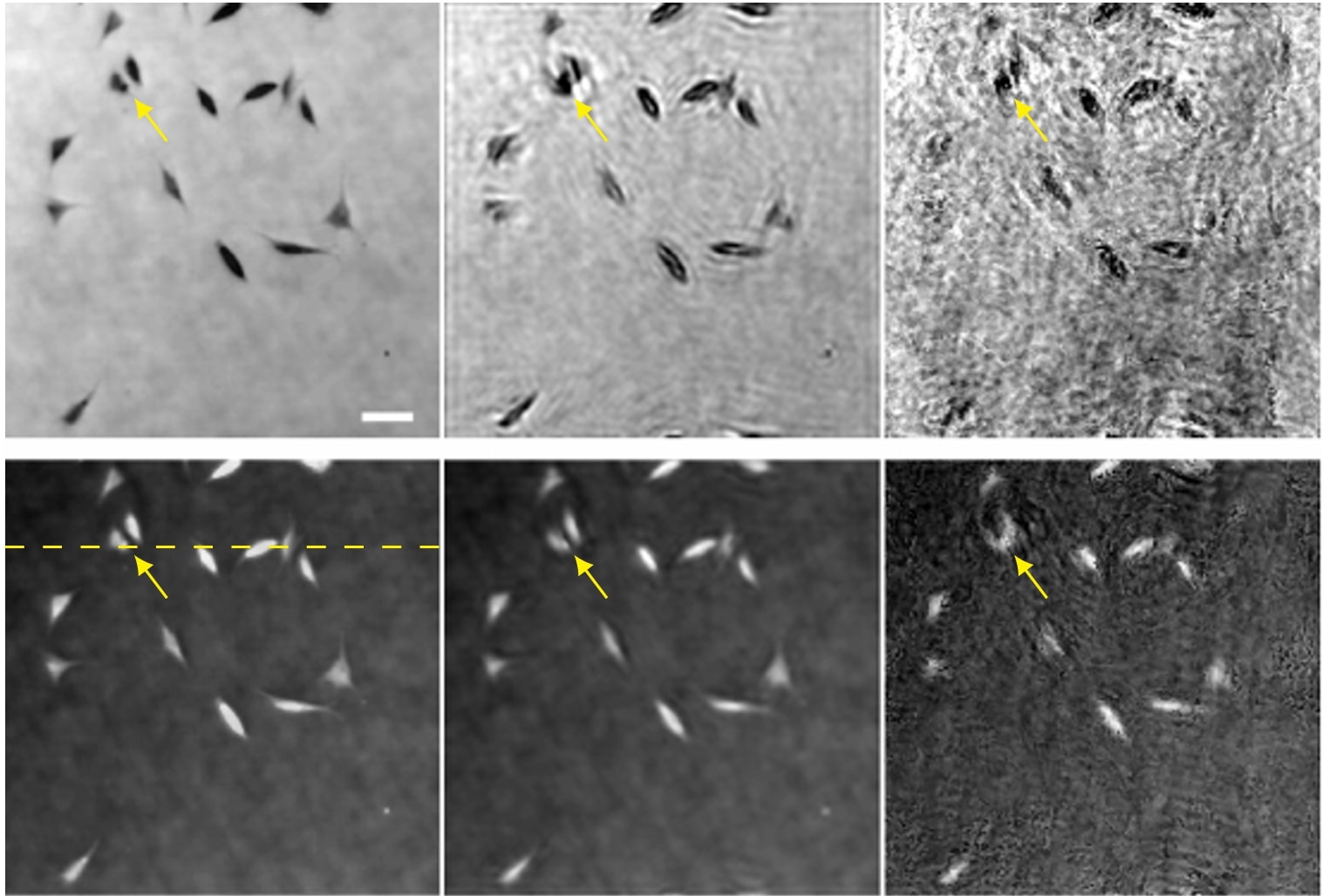
The goal of the project is to build new algorithms to reconstruct images in Fourier ptychographic microscopy. This imaging modality is used to observe biological cells using a LED array microscope. Several low-resolution images are acquired then combined into a high-resolution complex-valued image. This technique enables the recovery of quantitative phase of biological samples, with high resolution and large field-of-view. However, combining the low-resolution images remains a challenging computational task.
We want to develop a PDE/wavelets-inspired technique for this reconstruction problem. The high-resolution image will be represented by a family of basis functions (e.g. box-splines) whose coefficients will be estimated by an interative algorithm, mininizing a loss functional, quantifying the quality of the reconstruction. The difficulty is that this loss function is non-convex and may admit several bad local minimas. Likewise to optimization of neural networks, this leads to non-convex (non-linear) optimization. One way to mitigate this issue is to iteratively refine the basis functions and thus the target image. The idea being to first find the low-frequencies of the target image, then build your way up to high frequencies. This technique called multiresolution or multigrid, allows the algorithm to converge much faster and more reliably.
The task of the project is to build this multiresolution framework for Fourier Ptychography. You will be provided a Python library modelling the Fourier Ptychographic reconstruction on experimental images of biological cells, as well as a library for box-splines modeling. You will have to understand and bridge both codes into an efficient multiresolution reconstruction algorithm. The student should be comfortable in Python and numerical optimization.
- Supervisors
- Jonathan Dong, jonathan.dong@epfl.ch, EPFL
- Vincent Guillemet, vincent.guillemet@epfl.ch, EPFL