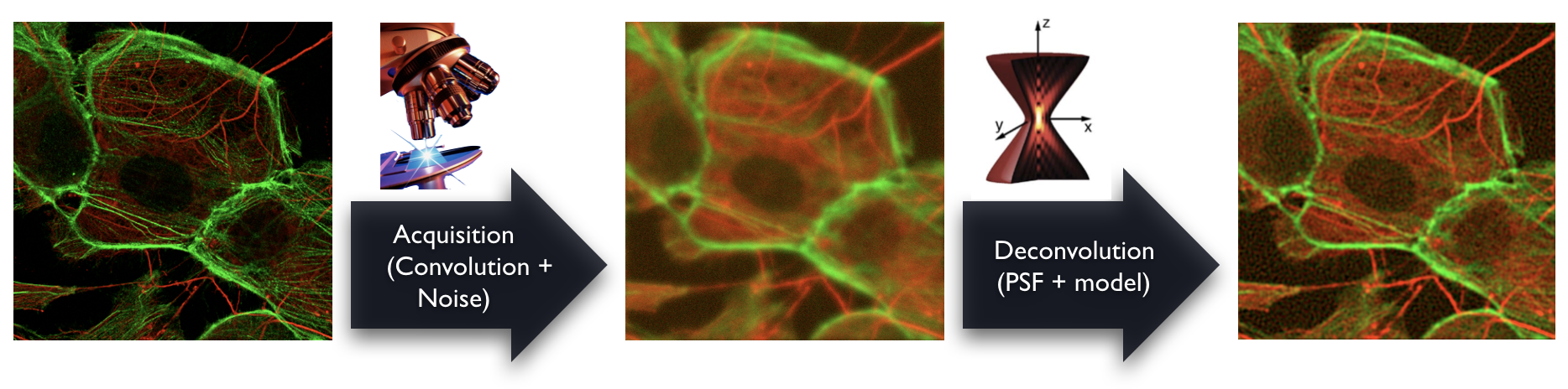
The deconvolution is an image-processing technique that restores the effective specimen representation for a 3D microscopy images. Various software packages for deconvolution are available, both commercial ones and open-source ones. They are computationally extensive requiring high-end processors and huge memory capacities. Despite the effort to provide user-friendly solutions, the deconvolution remains a challenging task in choosing the good software, the right algorithm and the correct settings.
Here, our contribution is to provide open software packages for 3D deconvolution microscopy, including a point-spread function (PSF) simulator, several 3D reference datasets and their corresponding PSF in order to compare deconvolution, and some benchmarks and comparison that we performed in the the past. In addition, we highlight advanced methods of deconvolution that we have developed in our laboratory.
The remasterized Java deconvolution tool • 2017
DeconvolutionLab2 is freely accessible and open-source; it can be linked to well-known imaging software platforms, ImageJ, Fiji, ICY, Matlab, or run as Java stand-alone software. DeconvolutionLab2 contains all the tools to perform 3D deconvolution on real images and real PSF, it also has modules to make simulation in order to validate algorithms. The backbone of this library that contains the number-crunching elements of the deconvolution task. It has a open architecture to easily plug deconvolution algorithms. The current list of built-in algorithms includes: Naive inverse filtering, Regularized inverse filtering, Landweber with positivity constraint, Tikhonov-Miller (ICTM), Fast iterative soft-thresholding (FISTA), Richardson-Lucy, Richardson-Lucy with total-variation regularization. The source code is written in Java 1.6, as close as possible to the text-book definition of the algorithms. Inquisitive minds inclined to peruse the code will find it fosters the understanding of deconvolution.
Reference
D. Sage, L. Donati, F. Soulez, D. Fortun, G. Schmit, A. Seitz, R. Guiet, C. Vonesch, M. Unser
DeconvolutionLab2: An Open-Source Software for Deconvolution Microscopy
Methods - Image Processing for Biologists, 115, 2017.
PDF
The original ImageJ deconvolution tool • 2007
DeconvolutionLab is an ImageJ plugin to deconvolve 3D images. DeconvolutionLab incorporates the most known algorithms of deconvolution with theirs parameters. Tuning these parameters could be difficult in deconvolution and may infer disappointing results. In DeconvolutionLab, these parameters can be choosen by the user or for some of them, they can be automicatilly estimated, which it is one of the main feature of DeconvolutionLab.
The Java software package to generate realistic 3D microscope Point-Spread Function
PSFGenerator is a software package that allows one to generate and visualize various 3D models of a microscope PSF. The current version has different models among them: Born & Wolf, Gibson & Lanni, and Richards & Wolf. PSF Generator is provided for several environments: as ImageJ/Fiji plugin, as an Icy plugin, and as a Java standalone application. The program requires only few parameters which are readily-available for microscopy practitioners.
Reference
H. Kirshner, F. Aguet, D. Sage, M. Unser
3-D PSF Fitting for Fluorescence Microscopy: Implementation and Localization Application
Journal of Microscopy 1, 2013.
Real specimen, multiple channels acquisition
This real dataset is composed of three stacks of images of a C. Elegans embryo. The deconvolution effects can be evaluated on different kinds of structures: extended objects (the chromosomes in the nuclei), filaments (the microtubules), and point-wise spots (a protein stained with CY3).
Real images and PSF, reference objects (donut) from Argolight
This real dataset of the 3D matrix of rings of an Argo-SIM slide from Argolight.
Result of deconvolution:
Download data, results of blind deconvolution, and the PSF [450 Mb]
This test volume contains a fluorescent bead of known dimension—its diameter is precisely 2.5 μm. This data have the advantage of offering a simple object on which it is easy to perform a quantitative validation of the recovering of shape and dimension, before and after deconvolution.
These synthetic data consist of six parallel hollow bars. The images have been blurred using a theoretical microscopic PSF, and corrupted by Gaussian noise and Poisson noise with several signal to noise ratios (SNR = 15, 30 dB). The knowledge of the ground-truth allows one to quantatively compare the deconvolution tools.
This datasets is a realistic-looking synthetic dataset representing a network of microtubules in a cell. It corresponds the GFP channel of the dataset used for the challenge on 3D deconvolution microscopy. The ground-truth, the PSF and some results are freely accessible upon a request by email.
Deconvolution - Making the most of fluorescence microscopy
Deconvolution is one of the most common image-reconstruction tasks that arise in 3D fluorescence microscopy. The aim of the challenges is to benchmark existing deconvolution algorithms and to stimulate the community to look for novel, global and practical approaches to this problem. It is primarily based on realistic-looking synthetic data sets representing various sub-cellular structures. In addition it relies on a number of common and advanced performance metrics to objectively assess the quality of the results.
Reference
C. Vonesch and S. Lefkimmiatis
Summary of the ISBI 2013 Grand Challenge on 3D Deconvolution Microscopy
IEEE International Symposium on Biomedical Imaging (ISBI'14), Beijing, China, 2014.
D. Sage, H. Kirshner, C. Vonesch, S. Lefkimmiatis, M. Unser
Benchmarking Image-Processing Algorithms for Biomicroscopy: Reference Datasets and Perspectives
Proceedings of EUSIPCO'13, Marrakech, Morocco, 2013.
Evaluation of 3D deconvolution software package
In modern optical microscopy and biological research deconvolution is becoming a fundamental processing step which allows for better image analysis. Deconvolution remains however a challenging task as the result depends strongly on the algorithm chosen, the parameters settings and the kinds of structures in the processed dataset. As a core facility of bio-imaging and microscopy, we aim with this study to compare the performances of different deconvolution software.
A. Griffa, N. Garin, D. Sage
Comparison of Deconvolution Software: A User Point of View—Part 1
—Part 2
G.I.T. Imaging & Microscopy 1, 2010.
Recently, the concept of sparsity has attracted a lot of interest for the resolution of inverse problems. The standard algorithm for solving the corresponding L1-regularized variational problem is known as the Thresholded Landweber (TL) algorithm. This MultiLevel Thresholded Landweber (MLTL) algorithm is an accelerated version of the TL algorithm that was specifically developped for deconvolution problems with a wavelet-domain regularization. By cycling through the wavelet-subbands in a multigrid-like fashion, it achieves a substantial speed-up.
C. Vonesch, M. Unser
A Fast Multilevel Algorithm for Wavelet-Regularized Image Restoration
IEEE Transactions on Image Processing, vol. 18, no. 3, March 2009.
Matlab implementation
Fast Multilevel Thresholded-Landweber Deconvolution Algorithm
Cédric Vonesch
Nonquadratic Hessian-based regularization methods can be effectively used for image restoration problems in a variational framework. Motivated by the great success of the total-variation (TV) functional, we extend it to also include second-order differential operators. Specifically, we derive second-order regularizers that involve matrix norms of the Hessian operator. The definition of these functionals is based on an alternative interpretation of TV that relies on mixed norms of directional derivatives. We show that the resulting regularizers retain some of the most favorable properties of TV, i.e., convexity, homogeneity, rotation, and translation invariance, while dealing effectively with the staircase effects.
S. Lefkimmiatis, J.P. Ward, M. Unser
Hessian Schatten-Norm Regularization for Linear Inverse Problems
IEEE Transactions on Image Processing, vol. 22, no. 5, May 2013.
S. Lefkimmiatis, A. Bourquard, M. Unser
Hessian-Based Norm Regularization for Image Restoration with Biomedical Applications
IEEE Transactions on Image Processing, vol. 21, no. 3, March 2012.
You'll be free to use this software for research purposes, but you must not transmit and distribute it without our consent. In addition, you undertake to include a citation whenever you present or publish results that are based on it. EPFL makes no warranties of any kind on this software and shall in no event be liable for damages of any kind in connection with the use and exploitation of this technology.
© 2021 EPFL • webmaster.big@epfl.ch • 28.07.2021